Lossless Compression Experiment
This page explains the general dynamics of using the enb.icompression.LosslessCompressionExperiment class.
This class is a subclass of enb.icompression.CompressionExperiment, which performs compression, decompression
and verifies that lossless compression is achieved.
Note
This example assumes you have understood the Basic workflow: enb.atable.ATable example, and that you have successfully followed the installation instructions in Installation.
Most ideas applicable to lossless compresesion experiments are also valid for lossy compression, described in the next page.
The
enb.icompressionandenb.isetsmodules implement most of the functionality specific to image compression experiments.
Lossless compression template installation
A template for lossless compression experiments is provided as the lossless-compression plugin in enb. To install this plugin under the .lc/ folder, run (after following Installation):
enb plugin install lossless-compression lc
You should see a message similar to the following:
Template 'lossless-compression' successfully installed into 'lc'.
You can check the full code and make your own modifications in lossless_compression_experiment.py under the installation folder.
Data curation
By default, all *.raw images under the enb.config.options.base_dataset_dir folder are considered as the input dataset. Images can be organized in subfolders as needed. Note that:
The dataset_files_extension can be changed from ‘raw’ to any other extension, or an empty string to recursively search for all files under the enb.config.options.base_dataset_dir folder.
The dataset_paths can be passed to the initializer of
enb.icompression.LosslessCompressionExperimentand other subclasses ofenb.icompression.CompressionExperiment. This overwrites the search under enb.config.options.base_dataset_dir.
Images used in this experiment need to satisfy the following requirements:
Be in raw (uncompressed, fixed-length output) format, preferably with ‘.raw’ extension. BSQ order is assumed in case more than one color component or spectral band is present.
Image file names must contain a tag such as ‘u8be-3x600x800’ in their name.
u or s should be used for unsigned or signed data
8, 16 or 32 indicate the bitdepth in bits of each sample
the geometry part of the tag is ZxYxX, where X, Y and Z are, respectively, the image’s width, height and number of spectral components (bands).
In the lossless-compression plugin, a sample image image_u8be-2x128x128.raw is included for ease of reference, and can be removed at any point.
Codec instantiation
Compression experiments use Codecs as enb.experiment.ExperimentTask subclasses. In particular, enb.icompression.LosslessCompressionExperiment
expects subclasses of enb.icompression.LosslessCodec.
The enb library provides a wide range of lossless and lossy codecs that can be installed via
the command line interface. A list of available codecs can be shown with the following command:
enb plugin list codec
It is recommended that you install your codec plugins under the ./plugins folder created under the installation dir of the lossless-compression plugin. In this example, we are going to install the zip codecs plugin as follows:
enb plugin install zip lc/plugins/zip
Once you have installed all codec plugins, a list of the codecs to be used in the experiment needs to be defined. In this example we will be using the following 4 codecs to compare BZIP2 and LZMA at their best and worst configuration levels
codecs = [plugins.zip.LZMA(compression_level=1),
plugins.zip.LZMA(compression_level=9),
plugins.zip.BZIP2(compression_level=1),
plugins.zip.BZIP2(compression_level=9)]
If we are happy with using all codecs installed in ./plugins with their default initializer, we can uncomment the following line of the template instead or in addition of defining our custom list of codecs
codecs += [cls() for cls in enb.misc.get_all_subclasses(enb.icompression.LosslessCodec)
if not "abstract" in cls.__name__.lower()]
Lossless experiment execution
The lossless_compression_experiment.py template is now ready to be run. To do so, simply invoke it with:
python ./lc/lossless_compression_experiment_example.py
This should produce the plots, analysis folders. Furthermore, a persistence_lossless_compression_experiment_example.py folder is created with persistence information, so that images do not need to be analyzed again, and that compression needs not be performed again unless you add any new codecs to your experiment.
If you check the code, you will see that the following two lines are responsible for the actual experiment execution
exp = LosslessExperiment(codecs=codecs)
df = exp.get_df()
where codecs is the list of codecs we defined above. The returned pandas.DataFrame df
can be used to analyze and plot the obtained results. This is automatically done in
the template for several columns of interest with the following code. You can check
the Result analysis and plotting with enb page for more information on how to analyze and plot
results.
scalar_columns = ["compression_ratio_dr",
"bpppc", "compression_time_seconds",
"decompression_time_seconds",
"compression_efficiency_1byte_entropy",
"compression_efficiency_2byte_entropy"]
column_pairs = [("bpppc", "compression_time_seconds"),
("compression_time_seconds",
"compression_efficiency_1byte_entropy"),
("compression_time_seconds",
"compression_efficiency_2byte_entropy")]
# Scalar column analysis
scalar_analyzer = enb.aanalysis.ScalarNumericAnalyzer(
# A scalar analysis summary is stored here
csv_support_path=os.path.join(options.analysis_dir,
"lossless_compression_analysis_scalar.csv"))
scalar_analyzer.get_df(
# Mandatory params
full_df=df, # the dataframe produced by exp
target_columns=scalar_columns, # the list of ATable column names
# Provide plotting hints based on the defined columns
column_to_properties=exp.joined_column_to_properties,
# Optional params
group_by="task_label", # one can group by any column name
selected_render_modes={"histogram"},
)
# Two column joint analysis
twoscalar_analyzer = enb.aanalysis.TwoNumericAnalyzer(
csv_support_path=os.path.join(
options.analysis_dir, "lossless_compression_analysis_twocolumn.csv"))
twoscalar_analyzer.get_df(
full_df=df,
target_columns=column_pairs,
column_to_properties=exp.joined_column_to_properties,
group_by="task_label",
selected_render_modes={"scatter"},
)
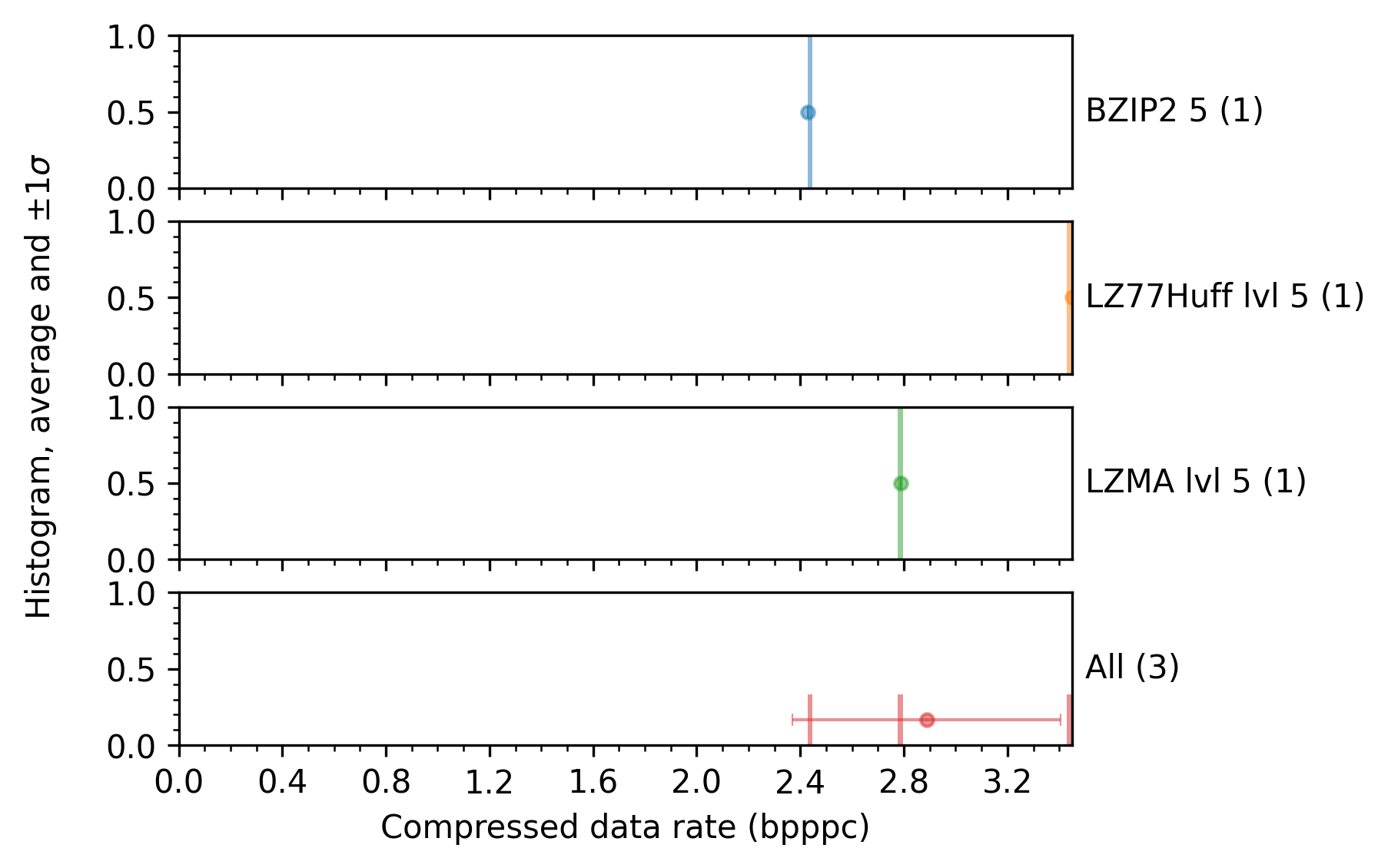
An example plot produced by this experiment, e.g., the compressed data rate in bits per sample, is shown in the next figure:
Saving the compressed and/or reconstructed files
One can easily copy all compressed and/or reconstructed files processed during the execution of a compression experiment. You simply need to add the compressed_copy_dir_path and/or the reconstructed_dir_path arguments to the constructor of your CompressionExperiment, e.g.:
exp = enb.icompression.LosslessExperiment(codecs=codecs,
compressed_copy_dir_path="compressed/",
reconstructed_dir_path="reconstructed/")
Note
Subclasses of LosslessCompressionExperiment automatically perform the lossless reconstruction verification, it is not needed to use the reconstructed_dir_path parameter and compare the reconstructed files manually.